Design
BioVisReport invokes specified plugins and biomedical data based on a user-defined Markdown file, and then generates an interactive website. This work is achieved through two components of BioVisReport, i.e., the website generator and the plugin system.
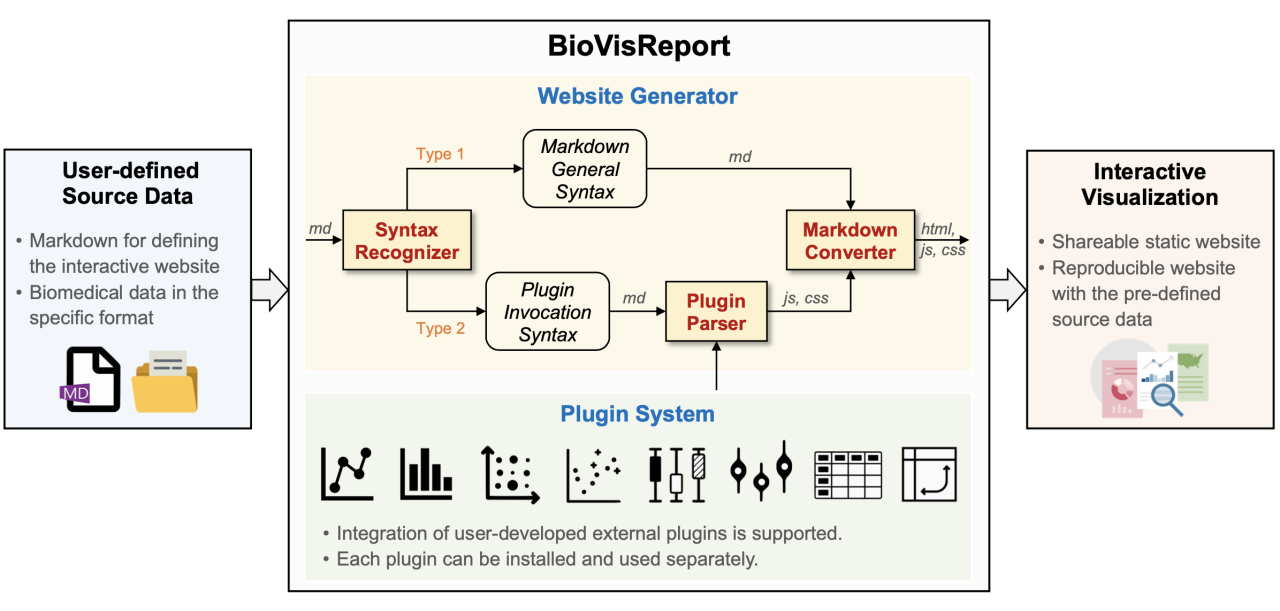
Website Generator¶
Website generator is built using Python and additional libraries including Jinja2, Markdown, Tornado, Mkdocs, and livereload. The generation of an interactive website requires the user to prepare data, set the website format and specify plot plugins. In BioVisReport, all the above tasks can be defined in a Markdown file. A syntax recognizer was developed to parse the plugin invocation syntax in the Markdown file and then execute these commands. Each plugin may receive one or more input files. The files can be located on a local disk or remotely, and can be in a tabular data file format (CSV or TSV), Rdata, or other formats. All data files are first downloaded or cached and then loaded using the built-in Python or R libraries. A Markdown converter receives the elements generated by the syntax recognizer and, at the same time, converts other commands in the Markdown file into the corresponding HTML statements to achieve the layout effect expected by the user.
Plugin System¶
The plugin system contains a variety of mutually independent plot plugins that can be invoked by the website generator. Our strategy is to develop separate plugins for each type of graph to achieve compatibility among multiple programming languages and a high level of extensibility. Each plugin has been encapsulated as a Python package and can be installed separately. BioVisReport recognizes Python packages through an API interface. This design strategy allows users with programming skills to develop customized plugins that are not yet supported in BioVisReport. All currently available plugins were developed using Python, JavaScript, R and additional libraries including Plotly, Dplyr, Shiny, ggplot2, pivotable, etc.